Following transcription, nascent pre-mRNAs undergo multiple processing steps, including Alternative Splicing (AS) and Alternative Polyadenylation (APA), to yield the final mature mRNA. Both AS and APA are ubiquitous mechanisms of gene expression that are tightly regulated in response to developmental and environmental stimuli. Different patterns of AS give rise to distinct proteins from a single gene, while APA typically regulates the level and duration of protein expression.
Our laboratory has traditionally focused on understanding how AS is regulated in T cells when they are stimulated by antigen during an immune response. We have identified >500 genes that undergo AS in response to T cell stimulation and have uncovered some of the molecular mechanisms and signaling pathways that lead to this regulation.
In our current efforts we are continuing to delineate the network of signaling events that lead to stimulation-induced AS in T cells and determine the molecular events that control AS in T cells at a detailed biochemical and biophysical level. We have also initiated studies into regulation of APA during T cell activation and are studying how AS impacts T cell function and differentiation. Finally, we are investigating additional aspects of how AS influences immune responses, including host-viral interactions and the role of AS in cancer and immunotherapy. Much of our current work can be summarized as addressing one of the four questions listed below. We are addressing these questions using a combination of biochemistry, genomics and computational approaches, biophysics and immunology.
1. Which Signaling Pathways lead to AS?
To identity which signaling pathways and which RNA-binding proteins (RBPs) control which AS events we are performing RNA-Seq of RNA collected following stimulation of T cells in the presence of chemical inhibitors of individual signaling pathways, or knock-down of individual RBPs, we have identified co-regulated AS events. Analysis of these co-regulated AS events allows us to infer common mechanisms of regulation through the use of computational and biochemical methods.
2. How do RBPs link Signaling to Splicing?
We are also using biochemical and molecular biology approaches to interrogate the mechanism and functional consequences of individual AS events of interest. For example, we recently demonstrated that the AS of the MKK7 gene is controlled by the JNK signaling pathway and the RBP CELF2, through the use of cell-based and biochemical methods. Moreover, this regulation serves to amplify JNK signaling through a positive feed-back loop (Martinez et al., Genes Dev 2015).
CELF2 is expressed in a JNK-dependent manner as shown by western blot (above left). This is attributed to JNK-dependent stabilization of CELF2 mRNA (above right). Future studies are aimed at determining this mechanism of regulated expression and stability of the CELF2 mRNA.
3. How is Activity of RBPs Regulated?
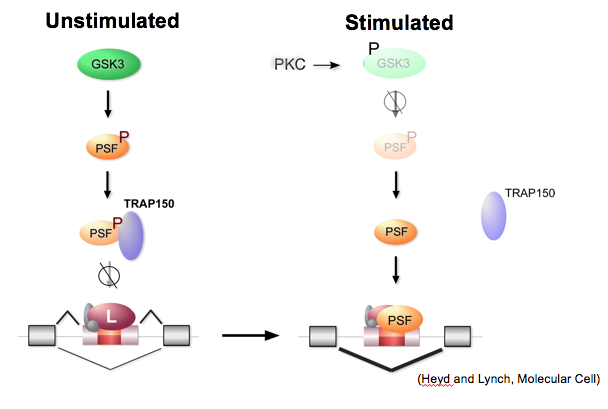
In studies of GSK3-PSF regulation of splicing we showed that phosphorylation of PSF by GSK3 controls the association of PSF with the protein TRAP150, which inhibits the binding of PSF to RNA.
More recently, we found that TRAP150 and RNA interact with an overlapping domain within PSF (Yarosh, et al., NAR 2015). Current studies are aimed at uncovering the structural of the PSF-TRAP and PSF-RNA interfaces using a range of biochemical and biophysical methods.
4. How does AS Shape Immune Response?
A. Immune system and cancer:
Cancers of lymphoid cells have been extensively linked to mis-regulation of splicing. In addition, we recently collaborated with the lab of Dr. Thomas-Tikhonenko (U Penn) and others to show that alternative splicing of CD19 allows B-ALL cells to develop resistance to anti-CD19 immunotherapy. Current projects in the lab involve continued investigation of the mechanism of alternative splicing in B-ALL, AML and other cancers.
B. Viral Infection
Another important immune response is that induced upon viral infection. In collaboration with the lab of Dr. Beatriz Fontoura (UT Southwestern) we have identified that two host RBPs, NS1-BP and hnRNP K, are required for proper splicing of the M transcript of influenza. We are now investigating this mechanism of regulation, and seeking to understand the role of AS in the host-viral interaction.